ADMET评价:预测和评价肠吸收HIA工作被RSC Advances接收
发表于 2017-05-25 10:05
Predicting human intestinal absorption with modified random forest approach: a comprehensive evaluation of molecular representation, unbalanced data, and applicability domain issues†
Ning-Ning Wang‡ a, Chen Huang‡b, Jie Donga, Zhi-Jiang Yaoac, Min-Feng Zhuac, Zhen-Ke Denga, Ben Lvc, Ai-Ping Lud, Alex F. Chenac and Dong-Sheng Cao *acd
*acd
aXiangya School of Pharmaceutical Sciences, Central South University, Changsha, 410013, P. R. China. E-mail: oriental-cds@163.com
bSchool of Mathematics and Statistics, Central South University, Changsha 410083, P. R. China
cThe 3rd Xiangya Hospital, Central South University, Changsha, 410000, P. R. China
dInstitute for Advancing Translational Medicine in Bone & Joint Diseases, School of Chinese Medicine, Hong Kong Baptist University, Hong Kong SAR, P. R. China
First published on 29th March 2017
Abstract
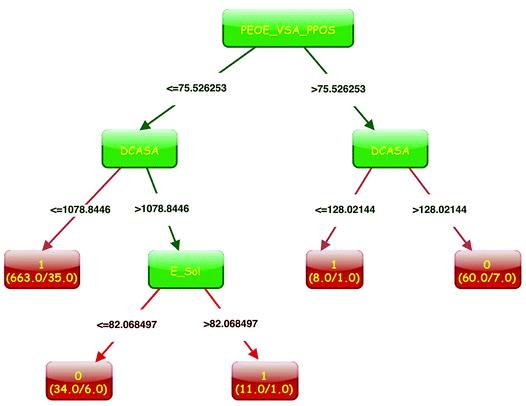
With the increase of complexity and risk in drug discovery processes, human intestinal absorption (HIA) prediction has become more and more important. Up to now, some predictive models have been constructed to estimate HIA of new drug-like compounds with acceptable accuracies, but there are still some issues to be explored including the limited and unbalanced HIA data, the performance of different types of descriptors and the application domain issues of published models. To address these problems, in this study, we collected a relatively large dataset consisting of 970 compounds, and 9 different types of descriptors were calculated for further modeling. For all the modeling processes, a parameter named samplesize in the random forest (RF) method was applied to balance the dataset. And then, classification models were established based on different training sets and different combinations of descriptors. After a series of modeling processes and various comparisons among these statistical results, we explored the aforementioned problems and evaluated the reliabilities of existing HIA classification models and subsequently obtained a robust and applicable model based on a combination of 2D, 3D, N+ and Nrule-of-five (for the training set, SE = 0.892, SP = 0.846; for the test set, SE = 0.877, SP = 0.813). Compared with other published models, our model exhibits some advantages in data size, model accuracy and model practicability to some extent. This structure–activity relationship model is necessary and useful for HIA prediction and it could be a convenient tool for virtual screening in the early stage of drug development.