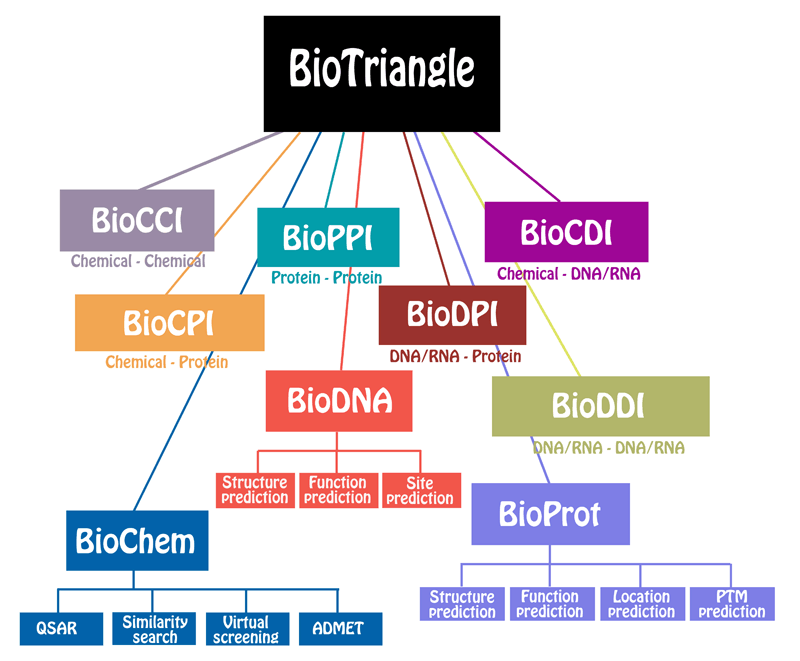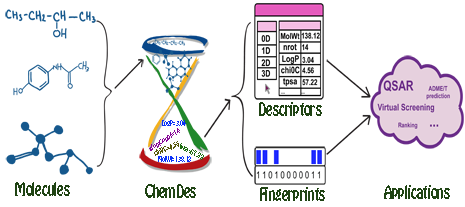
» ChemDes: An integrated web-based platform for molecular descriptor and fingerprint computation
ChemDes is a free web-based platform for the calculation of molecular descriptors and fingerprints, which provides more than 3,679 molecular descriptors that are divided into 61 logical blocks.In addition, it provides 59 types of molecular fingerprint systems for drug molecules, including topological fingerprints, electro-topological state (E-state) fingerprints, MACCS keys, FP4 keys, atom pairs fingerprints, topological torsion fingerprints and Morgan/circular fingerprints, et al.
» ProtrWeb: A web-based platform to calculate protein sequence-derived structural and physicochemical descriptors
Over the past two decades, statistical machine learning methods have been successfully employed in the structural, functional and interaction profiles research of proteins and peptides. The structural and physicochemical descriptors have been intensively applied in the research of protein structure and functionalities. ProtrWeb, a web server based on our R package protr, dedicated to compute such structural and physicochemical descriptors. Currently, ProtrWeb offers twelve types of the commonly used descriptors presented in the protr package.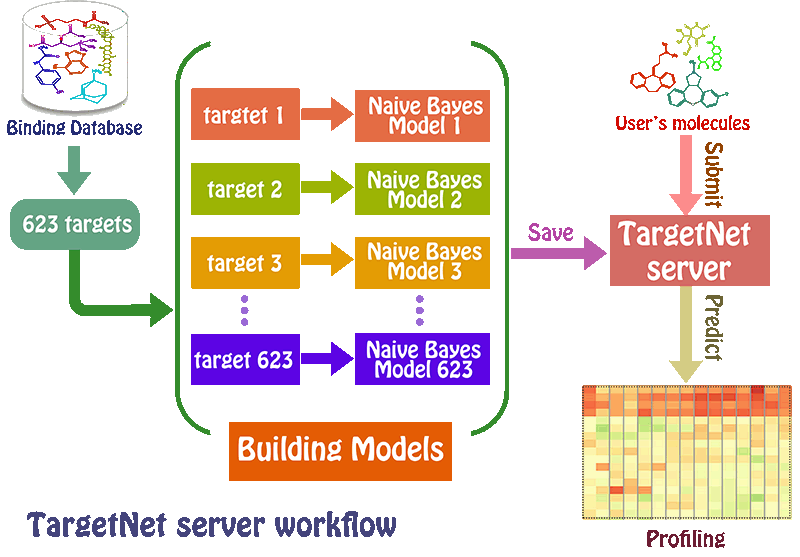
» TargetNet: A platform for predicting the binding of multiple targets for any given molecule
TargetNet is an open web server that could be used for netting or predicting the binding of multiple targets for any given molecule. TargetNet simultaneously constructs a large number of QSAR models based on current chemogenomics data to make future predictions. When the user submits a molecule, the server will predict the activity of the user’s molecule across 623 human proteins by establishing the high quality QSAR model for each human protein, thus generating a DTI profiling that can used as a feature vector for wide applications.